Research Article - Neuropsychiatry (2017) Volume 7, Issue 5
Tlr7 Deletion Selectively Ameliorates Spatial Learning but does not Influence beta Deposition and Inflammatory Response in an Alzheimers Disease Mouse Model
- Corresponding Author:
- Dr. Yi-Ping Hsueh
Institute of Molecular Biology, Academia Sinica
128, Academia Road, Section 2, Taipei 115
Taiwan, Republic of China.
Tel: +886-2-27899311
Fax: +886-2-27826085
Abstract
Objectives:
Alzheimer’s disease (AD) is highly associated with inflammation. Toll-like receptors (TLR), the critical pattern recognition receptors, initiate innate immune responses in a variety of cells. A role for TLR7 in AD has been postulated through its recognition of a specific miRNA that is upregulated in AD patients. In this report, we directly investigate the role of TLR7 in AD using mouse genetic models.
Methods:
5XFAD mice were used here as a mouse model for AD. Behavioral features, brain anatomy, amyloid beta (Aβ) deposition, microglial activation and inflammatory cytokine production of wild-type, 5XFAD, Tlr7–/Y, and Tlr7–/Y; 5XFAD mice were compared to evaluate the role of TLR7 in AD.
Results:
Open field and Barnes maze paradigms were used to assess the effect of Tlr7 knockout on behaviors of AD mice. Among the various behavioral features, the spatial learning performance in Barnes maze of 5XFAD mice was noticeably improved following Tlr7 knockout. Using immunostaining and quantitative real time-PCR (Q-PCR), our data indicated that the hallmark of AD brains-including Aβ deposition, activation of microglial cells and astrocytes, and upregulation of inflammatory cytokines-are not altered by Tlr7 deletion. These findings suggest that although Tlr7 is upregulated in 5XFAD mice and controls spatial learning of 5XFAD mice, TLR7 is not critical in the inflammatory responses of AD brains.
Conclusion:
Our results suggest the beneficial effect of Tlr7 deletion on the spatial learning process of 5XFAD mice, even though Aβ deposition and inflammation in the brains of 5XFAD mice are not ameliorated by Tlr7 deletion. The role of TLR7 differs from that of Tlr2 deletion in correcting AD pathological features. Thus, various TLRs likely carry out different functions and have differing or even opposite impacts on AD.
Keywords
5XFAD mice, Alzheimer’s disease, Cytokine, Microglia, Neurodegenerative disorder, Spatial learning, Toll-like receptor
Abbreviations
Aβ: Amyloid beta; AD: Alzheimer’s Disease; APP: Amyloid Precursor Proteins; GFAP: Glial Fibrillary Acidic Protein; IBA1: Ionized Calcium Binding Adaptor Molecule 1; Ifnβ: Interferon Beta; Il: Interleukin; PBS: Phosphate Buffered Saline; PFA: Paraformaldehyde; PSEN1: Presenilin 1; Q-PCR: Quantitative Real-Time Polymerase Chain Reaction; ssRNA: Single- Stranded RNA; TLR: Toll-Like Receptor
Introduction
Alzheimer’s disease (AD), a progressive neurodegenerative disorder, is tightly associated with neuroinflammation. Deposition of extracellular amyloid plaques, neurofibrillary tangles and microglial activation in the brain are the pathogenic hallmark of AD [1-8]. To initiate an inflammatory response, cells use pattern recognition receptors to recognize exogenous pathogen-associated molecular patterns, as well as endogenous damage-associated molecular patterns [9-11]. Toll-like receptors (TLR)-one type of pattern recognition receptors-comprise thirteen different members that recognize various pathogen-associated molecular patterns, including bacterial lipoproteins, bacterial and viral RNAs and DNAs, bacterial flagellin, zymosan and lipopolysaccharide [12,13]. TLRs also bind endogenous ligands [14]. Specifically, TLR2 can bind high-mobility group box 1 [15], as well as fibrillar Aβ peptides [16]. TLR2 activation of microglial cells by Aβ triggers neuroinflammation [16-18]. In addition, activation of TLR2, TLR4 and TLR9 increases Aβ clearance of BV2 microglia [19]. TLR3 and TLR7 recognize endogenous mRNA and miRNA [20-24]. Interestingly, miRNA let- 7 that is recognized by TLR7 is upregulated in the cerebrospinal fluid of patients with AD [23]. Intrathecal administration of let-7 induces Tlr7-dependent neurodegeneration in mice [23]. These studies suggest roles for TLRs in the neuroinflammatory responses of AD.
To further explore the role of TLRs in AD etiology, here, we investigated the effect of Tlr7 deletion on the phenotypes of 5XFAD mice, an AD mouse model expressing human Amyloid Precursor Proteins (APP) and Presenilin 1 (PSEN1) mutants [25]. We found that Tlr7 deletion did not influence microglial activation, inflammatory cytokine expression or Aβ deposition in 5XFAD mice, but it did ameliorate spatial learning of mutant mice in Barnes maze. These findings suggest that despite not influencing inflammatory responses in 5XFAD mice, Tlr7 deletion partially ameliorates the neural functioning of AD mice. These results imply that different players in inflammation participate differently in AD pathogenesis.
Materials and Methods
▪ Animals
Tlr7–/Y [26] and B6.Cg-Tg (APPSwFlLon, PSEN1* M146L* L286V) 6799Vas/J transgenic mice, also known as 5XFAD mice [25], on a C57BL/6 genetic background were purchased from the Jackson Laboratory. A breeding scheme using female 5XFAD was avoided to reduce the contribution of impaired maternal inflammatory responses to offspring behaviors. Thus, female Tlr7+/– mice were crossed with male 5XFAD mice to generate Tlr7–/Y; 5XFAD and TTlr7+/ Y;5XFAD (labeled as 5XFAD) male offspring for experiments. Mice were housed and bred in the animal facility of the Institute of Molecular Biology, Academia Sinica, under pathogen-free conditions and a 12 hr light/12 hr dark cycle with controlled temperature and humidity and free access to water and chow (LabDiet #5010). For behavioral analyses, mice were transferred to a separate behavior room with a 12 hr light/12 hr dark cycle control (lights off at 20:00) at least 1 week before behavioral assays. All animal experiments were performed with the approval of the Academia Sinica Institutional Animal Care and Utilization Committee and in strict accordance with its guidelines.
▪ Relative quantitative reverse transcription-polymerase chain reaction (Q-PCR)
Mouse whole brains were subjected to RNA extraction using Trizol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA), followed by DNase I (NEB) digestion for 30 min at 37°C to remove contaminating DNA. 5 μg RNA isolated from mouse tissues were then used for cDNA synthesis by the Transcriptor First Strand cDNA Synthesis Kit (Roche) with an oligo (dT)18 primer. A real-time PCR assay was performed using the LightCycler480 (Roche) and the Universal ProbeLibrary probes (UPL; Roche) system. The accession numbers of genes, primers and their paired probes, designed using the Assay Design Center Web Service (http://qpcr.probefinder. com/roche3.html), are as follows:
1. Tlr1: NM_030682.1, 5’-ctgagggtcctgataatgtcct-3’, 5’- tccagctctgtgttgaatttga-3’, #109.
2. Tlr2: NM_011905.3, 5’-GGGGCTTCACTTCTCTGCTT-3’, 5’- AGCATCCTCTGAGATTTGACG-3’, #50.
3. Tlr3: NM_126166.4, 5’-GATACAGGGATTGCACCCATA-3’, 5’- TCCCCCAAAGGAGTACATTAGA-3’, #26.
4. Tlr4: NM_021297.2, 5’-ggactctgatcatggcactg-3’, 5’-ctgatccatgcattggtaggt-3’, #2.
5. Tlr5: NM_016928.2, 5’-ctggagccgagtgaggtc-3’, 5’-cggcaagcattgttctcc-3’, #1.
5. Tlr5: NM_016928.2, 5’-ctggagccgagtgaggtc-3’, 5’-cggcaagcattgttctcc-3’, #1.
7. Tlr7: NM_133211.3, 5’-TGATCCTGGCCTATCTCTGAC-3’, 5’- CGTGTCCACATCGAAAACAC-3’, #25.
8. Tlr8: NM_133212.2, 5’-CAAACGTTTTACCTTCCTTTGTCT-3’, 5’-ATGGAAGATGGCACTGGTTC-3’, #56.
9. Tlr9: NM_031178.2, 5’-GAATCCTCCATCTCCCAACAT-3’, 5’- CCAGAGTCTCAGCCAGCACT-3’, #79.
10. Tlr11: NM_205819.2, 5’-atggggctttatcccttttg-3’, 5’-agatgttattgccactcaacca-3’, #1.
11. Tlr12: NM_205823.2, 5’-tctgaggggtaagggagaca-3’, 5’-gcagtgggacacgaatacatc-3’, #103.
12. Tlr13: NM_205820.1, 5’-acttggccggacagtgtt-3’, 5’-gcccaacgcatttctgat-3’, #66.
13. Il6: NM_031168.1, 5’-GCTACCAAACTGGATATAATCAGGA-3’, 5’-CCAGGTAGCTATGGTACTCCAGAA-3’, #6.
14. Il-1b: NM_008361.3, 5’-AGTTGACGGACCCCAAAAG-3’, 5’-AGCTGGATGCTCTCATCAGG-3’, #38.
15. Ifnb1: NM_010510, 5’-CACAGCCCTCTCCATCAACTA-3’, 5’-CATTTCCGAATGTTCGTCCT-3’, #78.
16. Tbp: NM_013684.3, 5’-ggcggtttggctaggttt-3’, 5’-gggttatcttcacacaccatga-3’, #107.
The PCR thermal profile was: denaturation at 95°C for 10 min; 45 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 30 sec, and extension at 72°C for 1 sec; and a final cooling step at 40°C for 30 sec.
▪ Hematoxylin and eosin staining
Mice were perfused sequentially with phosphate buffered saline (PBS) alone and 4% paraformaldehyde (PFA) in PBS, and then brains were dissected and post-fixed with 4% PFA/PBS overnight at 4°C. After fixation and cryopreservation with 30% sucrose, brains were embedded in OCT compound (Sakura, Tissue-Tek) and sliced into 50 μm-thick coronal sections using a cryostat. Brain sections were mounted on slides before being dehydrated sequentially with 70% EtOH and 95% EtOH. Sections were incubated in hematoxylin solution (Sigma) for 5-6 min. After washing, sections were then destained with acid (70% EtOH with 30 mM HCl) and rinsed with water and transferred to eosin solution (Sigma) for 1 min. After washing with water, the sections were destained with 70% EtOH and dehydrated for mounting. Images were acquired using an Axio imager M2 microscope (Carl Zeiss) with a 10x objective lens/NA 0.45 (Plan Apochromat; Carl Zeiss). The sizes of ventricles and brains were quantified using ImageJ (version 1.48).
▪ Immunohistochemistry and quantification
After behavioral assays, mouse brains were collected for immunohistochemical analysis. The brain sections were permeabilized with 0.5% Triton-X 100 in PBS for 10 min. After washing with PBS three times, slices were incubated with 70% formic acid for 20 min. The sections were then washed twice with PBS and blocked with blocking solution (1% bovine serum albumin, 3% horse serum and 0.3% Triton-X 100 in PBS) for 30 min. After blocking, sections were incubated individually with mouse monoclonal anti-beta amyloid antibody (1:200; Abcam, ab11132, DE2B4), rabbit polyclonal anti- IBA1 antibody (1:200; Wako, 019-019741) and mouse monoclonal anti-GFAP (1:200; Chemicon, MAB3402) in blocking solution overnight at 4°C. Sections were then incubated with Alexa Fluor-488- or Alexa Fluor-594- conjugated secondary antibodies (1:500) for 2 hr at room temperature and counter-stained with DAPI. The slices were then mounted with Vectashield mounting medium (H-1000; Vector Laboratories) and visualized at room temperature with an Axio imager M2 microscope (Carl Zeiss) equipped with a 10x objective lens/NA 0.45 (Plan Apochromat; Carl Zeiss). Immunofluorescence images were captured with a digital camera (Rolera EM-C2) driven by the digital image processing software Zen Blue (Carl Zeiss). The total areas of Aβ, IBA1 and GFAP immunoreactivities were quantified using ImageJ (version 1.48), as described [27-29]. Briefly, all images were converted into 8-bit and adjusted with auto-threshold (“Moments” and “B&W” modes in “Dark background”). The area of immunoreactivity was analyzed using “Analyze Particles” with the setting of 30-infinity pixel units and 0.01-1.00 circularity. The quantitatives results were manually corrected to remove artifacts, such as the ventricle and the edges of brain sections. The hippocampus or cortex was outlined using polygon selection. The data were then transferred to Excel to calculate the ratio of area of each antigen to total area of hippocampus or cerebral cortex.
▪ Behaviors
Eight-month-old male mice were analyzed sequentially with the open field and Barnes maze paradigms with 1-2 week intervals. The open field test was carried out during 16:00~18:00 and performed as described previously [27]. Briefly, mice were placed in the center of a transparent plastic box (40 × 40 × 30 cm) for free exploration for 10 min. Mouse behavior was recorded by camera from above. The total moving distance and the time spent in the corners and center area were quantified using the Smart Video Tracking system (Panlab, Barcelona, Spain). Grooming and rearing were manually counted. For Barnes maze, the test was carried out during 13:00~18:00. Mice were individually placed in a cylindrical start chamber in the middle of a maze, which constituted a white circular platform (100 cm in diameter) with 40 equally-spaced holes around the perimeter. At day 0, mice were gently guided to enter the target hole by hand after removing the start chamber. Once mice entered the hole, the target hole was covered and mice were allowed to stay in it for 2 min. Over the following four days (the training phase), mice received 4 trials per day, with an inter-trial interval of 15 min for a total of 4 days. For each trial, mice were allowed to freely explore the maze until they entered the target hole. Mice were camera-recorded for 180 sec per trial from above. If mice had not reached the target hole within 180 sec, mice were guided by the observer to enter the target hole. The duration was recorded as 180 sec. The duration from the start point to enter the target hole was measured as the escape latency. The differences between the first trial and trials 2, 3 and 4 of day 1 were used to indicate escape latency savings.
▪ Statistical Analysis
Statistical analyses were performed using an unpaired Student’s t test by GraphPad Prism (GraphPad Software, La Jolla, CA) and two-way ANOVA with Bonferroni’s multiple comparison by SigmaStat 3.5 (Systat Software, Inc., San Jose, California). Specific methods for each data analysis are indicated in figure legends. Data are presented as the mean plus SEM (n > 5) or mean plus SD (n < 5).
Results
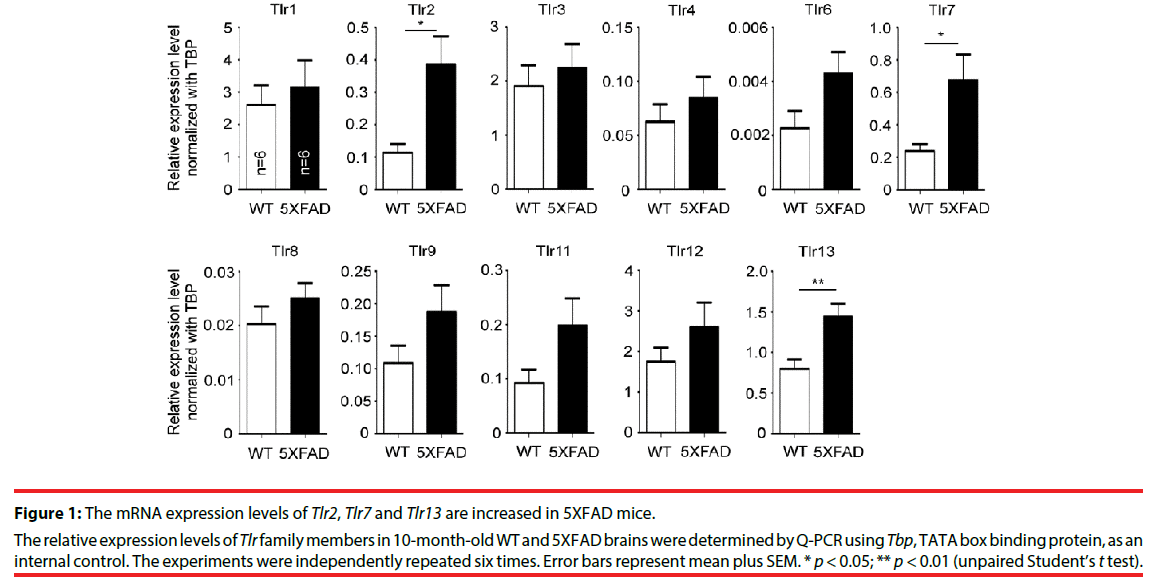
▪ Expression of Tlr2, Tlr7 and Tlr13 is increased in 5XFAD brains
To study the role of TLRs in AD, the expression levels of various TLRs in 5XFAD mice were examined. Ten-month-old mouse brains were analyzed by quantitative RT-PCR (Q-PCR). In general, the mRNA expression levels of TLRs tended to be higher in 5XFAD brains compared with wild-type littermates, but only Tlr2, Tlr7 and Tlr13 were significantly upregulated (Figure 1, for Tlr2, P = 0.0140; for Tlr7, P = 0.0251; for Tlr13, P = 0.0085). TLR2 has previously been shown to directly interact with Aβ and promote cell uptake of Aβ peptide to activate microglial cells [16,17,19,28]. TLR13 specifically recognizes bacterial ribosomal single-stranded RNA [29-31]. The role of TLR13 in recognition of endogenous dangerous signals is still unclear. TLR7 recognizes Let-7 miRNA that is increased in the cerebrospinal fluid of AD patients [23], but how TLR7 is involved in AD is unclear. Here, we explored the relevance of TLR7 in AD pathogenesis using 5XFAD and Tlr7 knockout mice as models.
Figure 1: The mRNA expression levels of Tlr2, Tlr7 and Tlr13 are increased in 5XFAD mice.
The relative expression levels of Tlr family members in 10-month-old WT and 5XFAD brains were determined by Q-PCR using Tbp, TATA box binding protein, as an internal control. The experiments were independently repeated six times. Error bars represent mean plus SEM. * p < 0.05; ** p < 0.01 (unpaired Student’s t test).
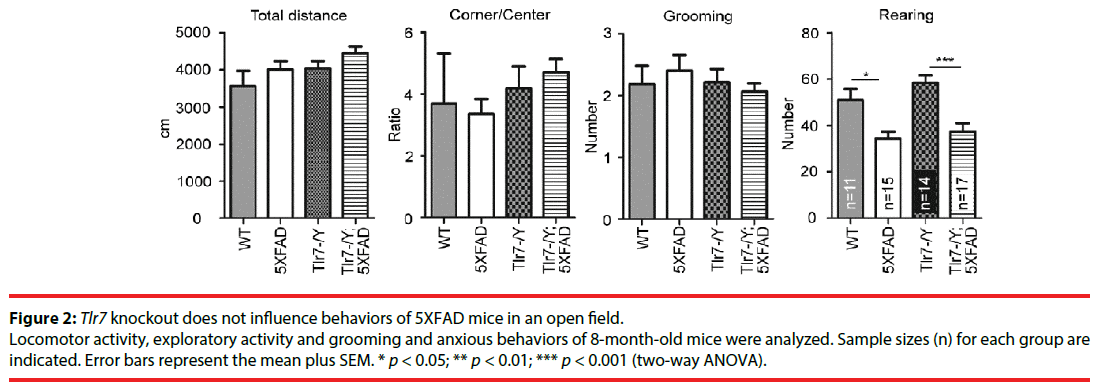
Tlr7 deletion does not noticeably alter behaviors of 5XFAD mice in open field
We investigated the effect of Tlr7 deletion on behaviors of 5XFAD mice. Wild-type, 5XFAD, Tlr7–/Y and Tlr7–/Y;5XFAD mice were subjected to behavioral analyses starting at 8 months. Deletion of Tlr7 in Tlr7–/Y mice had been validated in previous studies [24,32]. In open field, we assessed the locomotor, anxious, repetitive, and exploratory activities of mice. For travel distance, the ratio of corner to center, or the number of grooming events in the open field, there was no difference among the different mouse lines (Figure 2), suggesting that locomotion, anxiety and repetitive behaviors of mice were not affected by Tlr7 knockout or expression of APP and PSEN mutants. We noticed that 5XFAD mice had lower numbers of rearing events compared to wild-type mice (Figure 2). Tlr7 knockout did not ameliorate the limited rearing behavior of 5XFAD mice (Figure 2). Thus, our data suggest that Tlr7 knockouts does not alter the behavioral features of 5XFAD mice in an open field, including for locomotion, anxiety, exploration or repetitive behaviors.
Figure 2: Tlr7 knockout does not influence behaviors of 5XFAD mice in an open field.
Locomotor activity, exploratory activity and grooming and anxious behaviors of 8-month-old mice were analyzed. Sample sizes (n) for each group are indicated. Error bars represent the mean plus SEM. * p < 0.05; ** p < 0.01; *** p < 0.001 (two-way ANOVA).
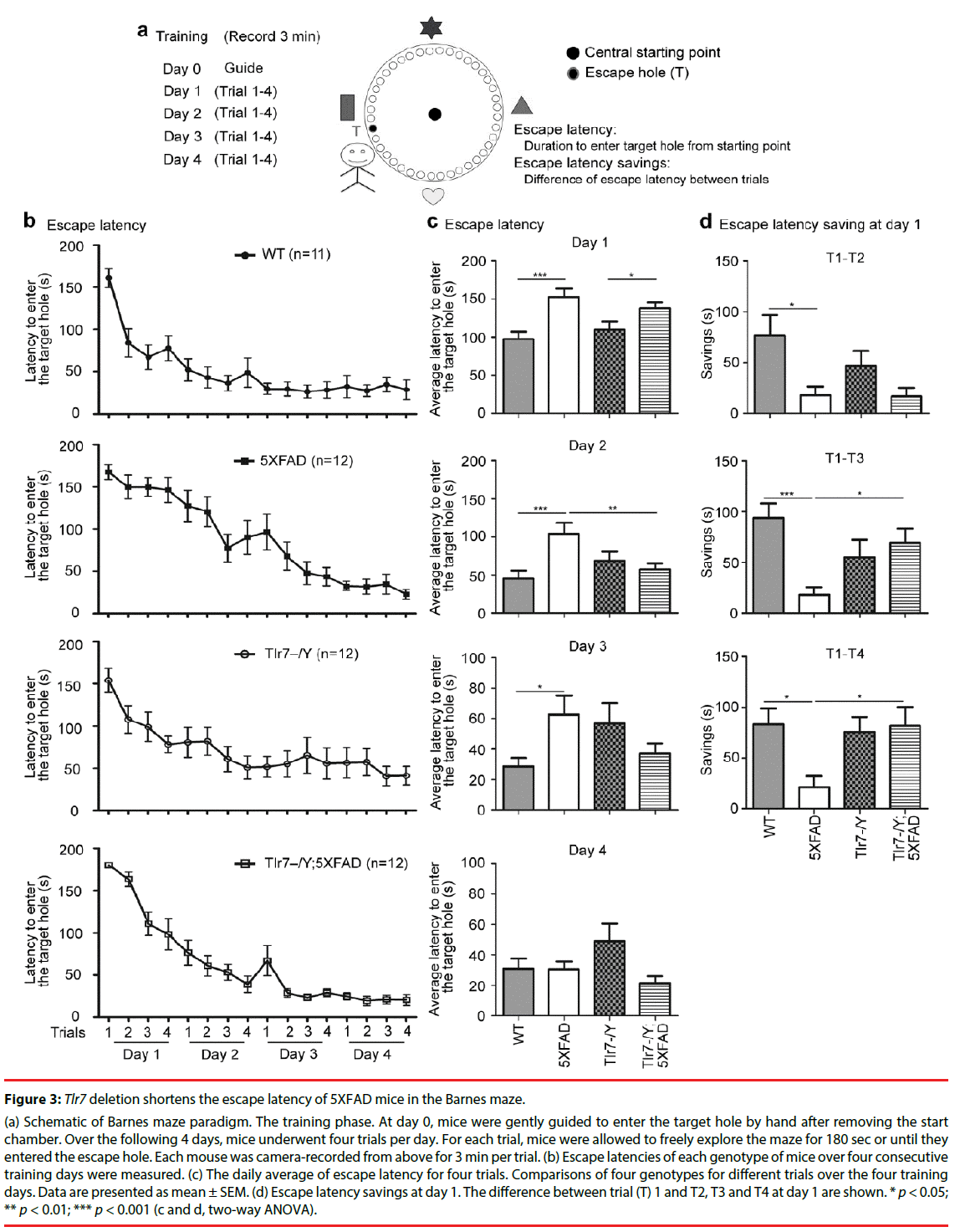
Tlr7 knockout ameliorates spatial learning of 5XFAD mice
We then used Barnes maze to analyze spatial learning of the mutant mice. Mice were subjected to four consecutive training days (days 1 to 4). Four trials were conducted in each training day. Two parameters-including escape latencies and escape latency savings between trials on day 1-were used to assess spatial learning in Barnes maze (Figure 3). Escape latency is defined by the latency to enter the target escape hole. During training, mice became quicker at entering the target escape hole, and these escape latency savings, which indicate the differences in escape latencies between trials, reflect how fast mice learn to find and enter the target escape hole (Figure 3a).
Figure 3: Tlr7 deletion shortens the escape latency of 5XFAD mice in the Barnes maze.
(a) Schematic of Barnes maze paradigm. The training phase. At day 0, mice were gently guided to enter the target hole by hand after removing the start chamber. Over the following 4 days, mice underwent four trials per day. For each trial, mice were allowed to freely explore the maze for 180 sec or until they entered the escape hole. Each mouse was camera-recorded from above for 3 min per trial. (b) Escape latencies of each genotype of mice over four consecutive training days were measured. (c) The daily average of escape latency for four trials. Comparisons of four genotypes for different trials over the four training days. Data are presented as mean ± SEM. (d) Escape latency savings at day 1. The difference between trial (T) 1 and T2, T3 and T4 at day 1 are shown. * p < 0.05; ** p < 0.01; *** p < 0.001 (c and d, two-way ANOVA).
Escape latencies of wild-type, 5XFAD, Tlr7–/Y and Tlr7–/Y; 5XFAD mice at 9 months were analyzed. Among the four analyzed mouse lines, 5XFAD mice had the slowest learning curve, as their escape latency was slowly reduced during the training process compared to the other three mouse lines (Figure 3b). We averaged escape latencies of four trials for each training day (Figure 3c). At day 1, the average escape latencies of wildtype and Tlr7–/Y mice were shorter than those of 5XFAD and Tlr7–/Y; 5XFAD mice (Figure 3c). These results echo previous observations that spatial working memory is impaired in 5XFAD mice [27-35]. Tlr7 knockout did not noticeably ameliorate learning defects of 5XFAD mice at day 1 (Figure 3c). However, at day 2, the learning performance of Tlr7–/Y;5XFAD mice was improved and comparable to wild-type and Tlr7–/Y mice (Figure 3c). The rescue effect of Tlr7 deletion on 5XFAD mice was also reflected in escape latency savings. The escape latency savings for T1–T2, T1–T3 and T1–T4 on day 1 were much higher in wild-type mice compared to 5XFAD mice (Figure 3d). For Tlr7–/Y;5XFAD mice, only T1–T2 was comparable to 5XFAD mice. T1–T3 and T1–T4 escape latency savings of Tlr7–/Y;5XFAD mice were larger than those of 5XFAD mice (Figure 3d). These analyses suggest that Tlr7 deletion ameliorates spatial learning of 5XFAD mice in Barnes maze.
Tlr7 deletion does not influence Aβ deposition and inflammatory responses in 5XFAD mice
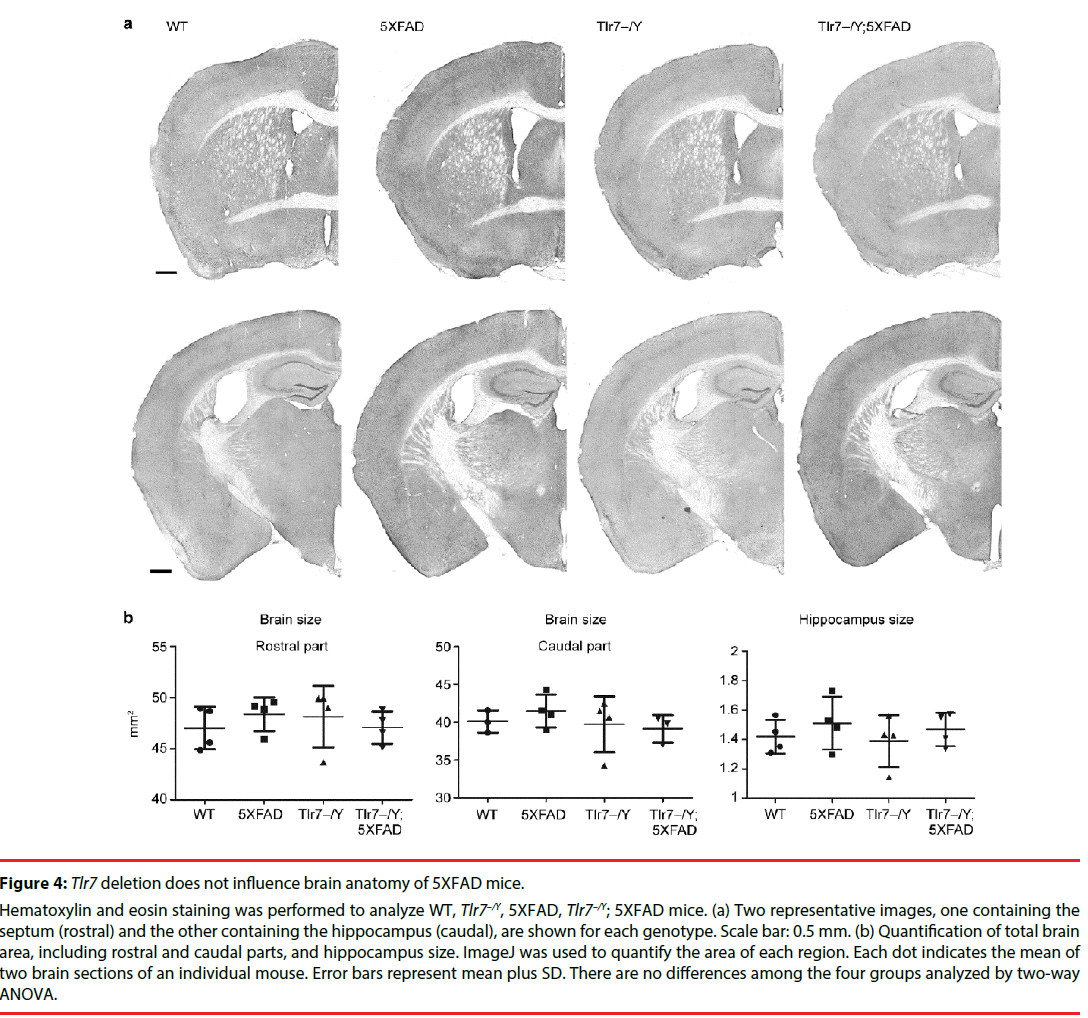
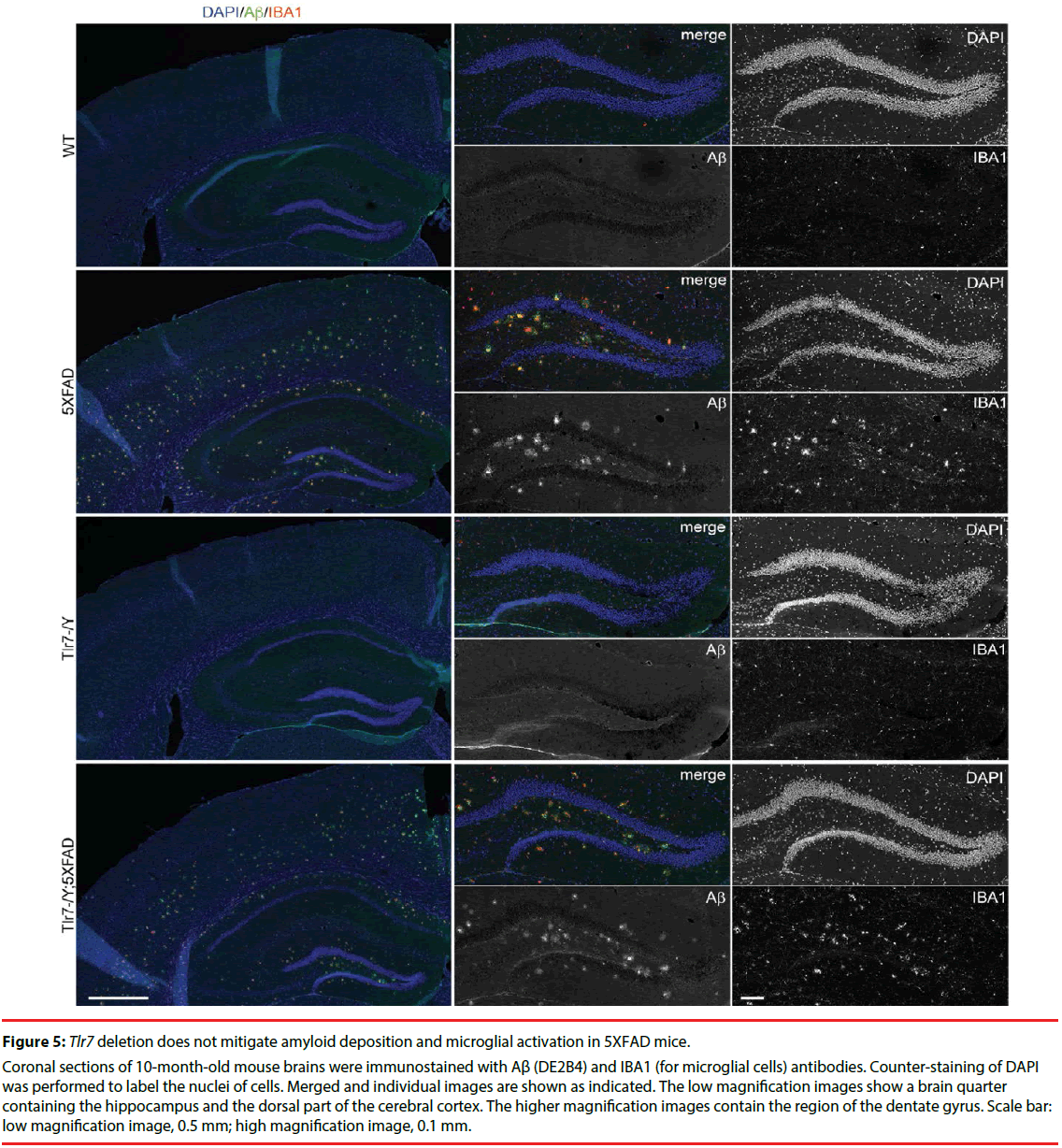
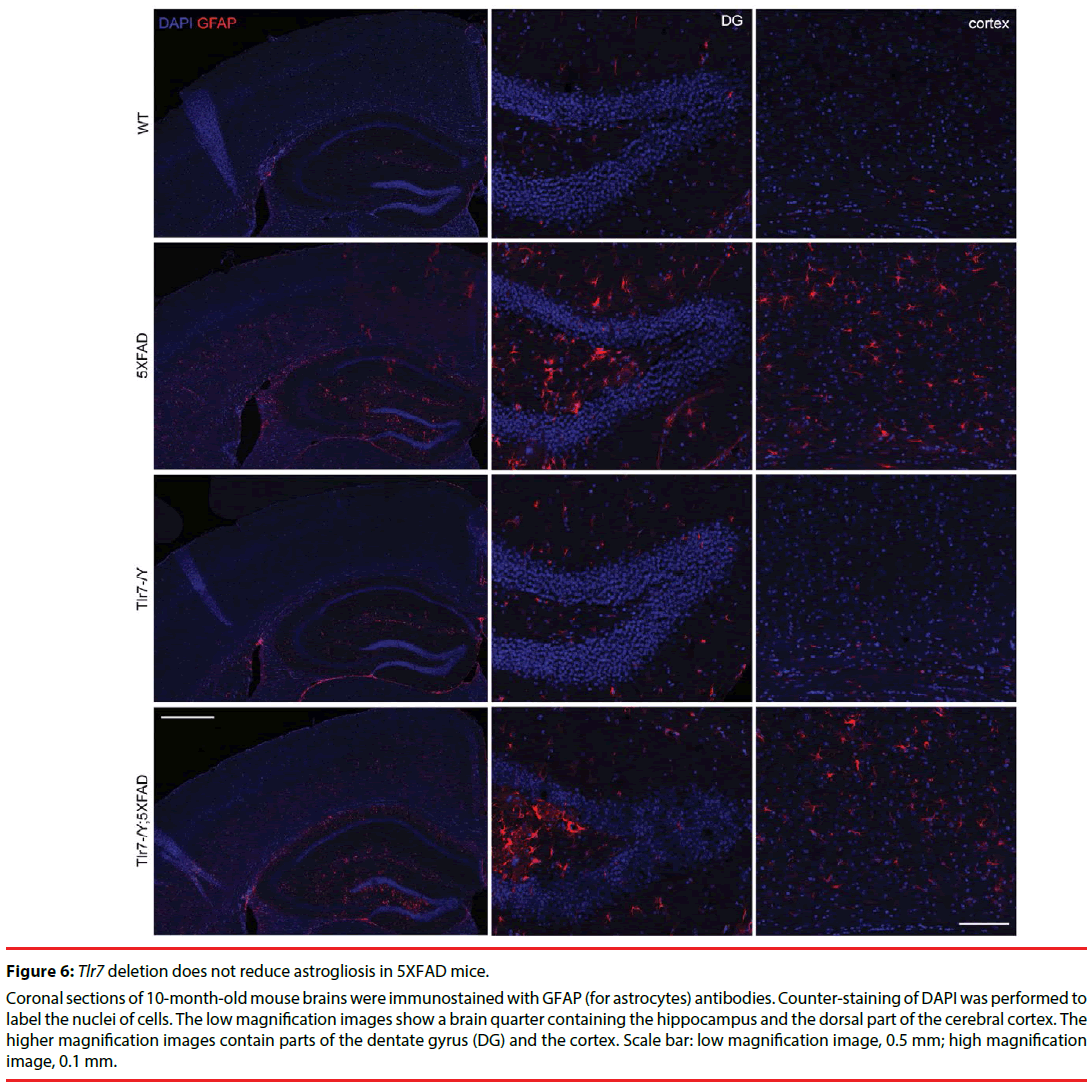
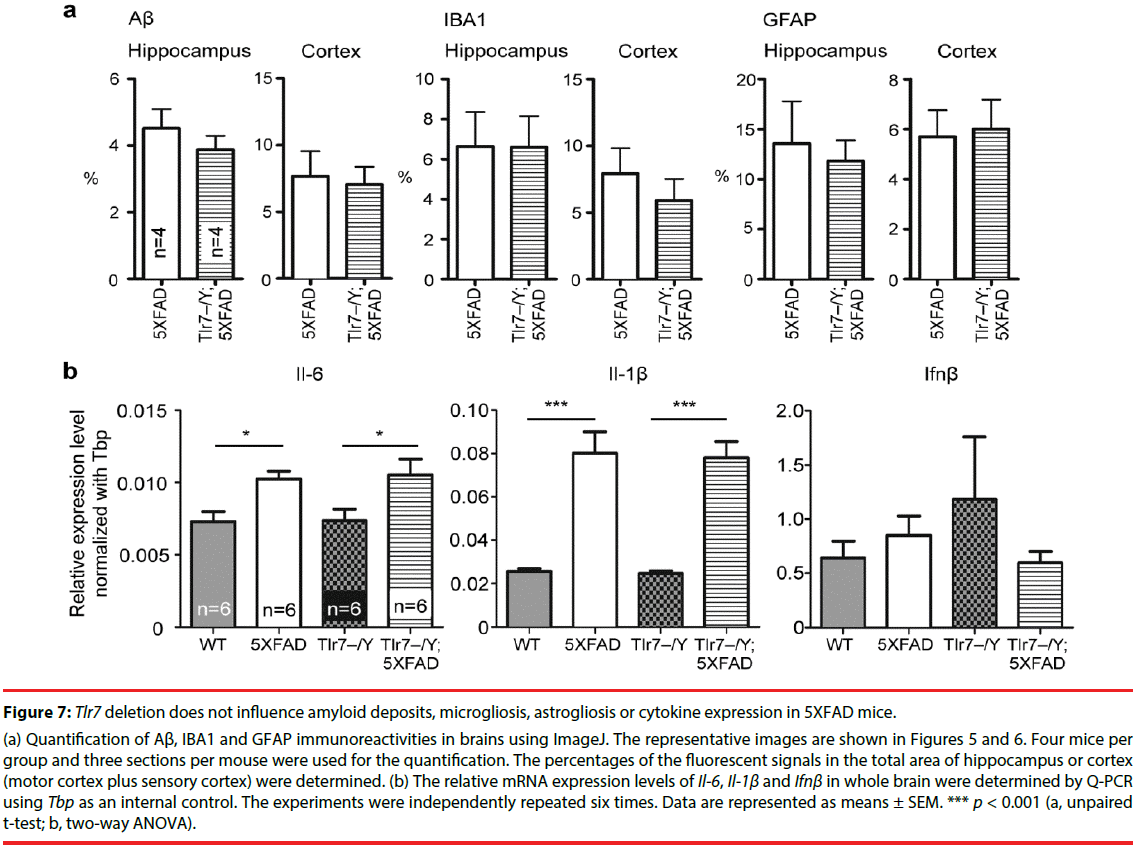
We then investigated whether Tlr7 deletion reduces the inflammatory response in the brains and thus influences mouse performance in Barnes maze. HE staining was first performed to examine the anatomical features of mouse brains. In general, deletion of Tlr7 did not noticeably influence the brain anatomy of 5XFAD mice, as the morphologies of brain sections prepared from WT, Tlr7–/Y, 5XFAD and Tlr7–/Y; 5XFAD were comparable. There were no differences in the sizes of the brains and hippocampi among these four genotypes of mice (Figure 4). A previous study had shown that, in 5XFAD brains, Aβ deposition begins at ~6 week of age and reached a plateau at ~9-12 months of age [25]. Using immunofluorescence staining, we then monitored Aβ deposition in 10-monthold mice. We found that both 5XFAD and Tlr7–/Y; 5XFAD mice had prominent Aβ plaques in brains (Figure 5). Consistent with Aβ deposition, profound immunoreactivities of Ionized calcium Binding Adaptor molecule 1 (IBA1), a microglia marker, and Glial Fibrillary Acidic Protein (GFAP), an astrocyte marker, were also found in 5XFAD and Tlr7–/Y; 5XFAD mice, but these immunoreactivities were very weak in wild-type and Tlr7–/Y mice (Figures 5 and 6). We then quantified Aβ deposition and activation of microglial cells and astrocytes. The results showed that total areas of Aβ plaques in mouse cerebral cortices and hippocampi were not altered by Tlr7 deletion (Figure 7a). Similarly, microglial activation, as indicated by immunostaining using IBA1 antibody, was not influenced by Tlr7 deletion (Figure 7a). In addition, astrocyte activation in the brains, as revealed by GFAP staining, was not obviously different between Tlr7–/Y; 5XFAD and 5XFAD mice (Figure 7a). These results suggest that Aβ clearance and activation of microglial cells and astrocytes in 5XFAD mice were not altered by Tlr7 deletion.
Figure 4: Tlr7 deletion does not influence brain anatomy of 5XFAD mice.
Hematoxylin and eosin staining was performed to analyze WT, Tlr7–/Y, 5XFAD, Tlr7–/Y; 5XFAD mice. (a) Two representative images, one containing the septum (rostral) and the other containing the hippocampus (caudal), are shown for each genotype. Scale bar: 0.5 mm. (b) Quantification of total brain area, including rostral and caudal parts, and hippocampus size. ImageJ was used to quantify the area of each region. Each dot indicates the mean of two brain sections of an individual mouse. Error bars represent mean plus SD. There are no differences among the four groups analyzed by two-way ANOVA.
Figure 5: Tlr7 deletion does not mitigate amyloid deposition and microglial activation in 5XFAD mice.
Coronal sections of 10-month-old mouse brains were immunostained with Aβ (DE2B4) and IBA1 (for microglial cells) antibodies. Counter-staining of DAPI was performed to label the nuclei of cells. Merged and individual images are shown as indicated. The low magnification images show a brain quarter containing the hippocampus and the dorsal part of the cerebral cortex. The higher magnification images contain the region of the dentate gyrus. Scale bar: low magnification image, 0.5 mm; high magnification image, 0.1 mm.
Figure 6: Tlr7 deletion does not reduce astrogliosis in 5XFAD mice.
Coronal sections of 10-month-old mouse brains were immunostained with GFAP (for astrocytes) antibodies. Counter-staining of DAPI was performed to label the nuclei of cells. The low magnification images show a brain quarter containing the hippocampus and the dorsal part of the cerebral cortex. The higher magnification images contain parts of the dentate gyrus (DG) and the cortex. Scale bar: low magnification image, 0.5 mm; high magnification image, 0.1 mm.
We then investigated whether Tlr7 deletion alters cytokine expression levels in 5XFAD mouse brains. The mRNA levels of Il-6, Il- 1β and Ifnβ were determined by Q-PCR. We found that Ifnβ levels did not differ among the four genotypes (Figure 7b). Both Il-6 and Il- 1β were more highly expressed in 5XFAD and Tlr7–/Y; 5XFAD brains compared with wild-type and Tlr7–/Y mice (Figure 7b). When 5XFAD and Tlr7–/Y; 5XFAD mice were compared, the levels of Il-6 and Il-1β were also similar to each other (Figure 7b). These results echo those of a previous study showing that an inflammatory response is triggered by AD [33]. However, Tlr7 deletion did not influence cytokine expression in 5XFAD mice. For Tlr7–/Y mice, Il-6, Il-1β and Ifnβ expression levels were not noticeably different from those of WT mice (Figure 7b). These analyses suggest that Tlr7 deletion has no obvious effect on cytokine expression in either wild-type or 5XFAD mouse brains.
Figure 7: Tlr7 deletion does not influence amyloid deposits, microgliosis, astrogliosis or cytokine expression in 5XFAD mice.
(a) Quantification of Aβ, IBA1 and GFAP immunoreactivities in brains using ImageJ. The representative images are shown in Figures 5 and 6. Four mice per group and three sections per mouse were used for the quantification. The percentages of the fluorescent signals in the total area of hippocampus or cortex (motor cortex plus sensory cortex) were determined. (b) The relative mRNA expression levels of Il-6, Il-1β and Ifnβ in whole brain were determined by Q-PCR using Tbp as an internal contro
Discussion
In this report, we used knockout mice to explore the contribution of TLR7 to AD. Although TLR7 is one of the critical pattern recognition receptors that initiate inflammation, our analyses of glial cell numbers and cytokine expression suggest that TLR7 deletion does not noticeably influence inflammatory responses in AD mouse brains. TLR7 recognizes single-stranded RNA (ssRNA), including miRNA. The expression level of let-7, a miRNA that is recognized by TLR7, is elevated in AD patients [23]. Activation of TLR7 by adding synthetic let-7 miRNA induces neuronal cell death in both neuronal cultures and mouse brains [23,34]. Therefore, involvement of TLR7 in the neurodegeneration caused by AD has been postulated [23]. In our study, Tlr7 knockout does not noticeably alter anatomic and inflammatory features of 5XFAD brains. Our data do not suggest a role for TLR7 in AD-related inflammation in 5XFAD mice. Several possibilities may explain these conflicts. First of all, the responses of neurons and brains to acute treatment of a high dose of let-7 [23,24] may be stronger than the chronic reaction in AD brains. Thus, it may not be easy to observe the effect of TLR7 in 5XFAD mice. Second,in addition to TLR7, TLR8 also recognizes ssRNA. Our previous study indicated specific upregulation of Tlr8 expression in neuronal cultures and the brain, but not in spleen , of Tlr7 deficient mice. [36]. Upregulation of Tlr8 may compensate for the effect of Tlr7 knockout on inflammatory responses in 5XFAD brains. It should be investigated in the future whether Tlr8 is still upregulated in 10-month-old mice, particularly in the background of 5XAD. Finally, although let-7 is upregulated in AD patients, the cause of this upregulation is unclear. In our study, 8-10 month-old 5XFAD mice were used as the AD model. Perhaps, the expression level of let-7 in 5XFAD brains is not high enough to activate TLR7 under this condition. It is also interesting to investigate in the future whether let-7 is increased in 5XFAD mice.
Although our data do not suggest that TLR7 is critical for the inflammatory response in 5XFAD mice, we found that Tlr7 knockout noticeably ameliorates the spatial learning process of 5XFAD mice. Given our data on microglial activation and cytokine expression, these results suggest that the beneficial effect of TLR7 on AD mice is unlikely to be due to amelioration of the inflammatory response of microglia. Our previous studies have shown that neuronally-expressed TLR7 acts cell-autonomously to restrict dendritic growth of neurons in both neuronal cultures and brains [24,32,35]. Knockout or knockdown of Tlr7 promotes dendritic growth of hippocampal CA1 or layer 2/3 cortical neurons before three weeks of age [24,32], suggesting an important role of TLR7 in developmental stages. Thus, in Tlr7–/Y; 5XFAD mice, the influence of Tlr7 deletion on learning performance might be caused by a developmental effect. It would be intriguing to further investigate how TLR7 influences learning processes. Knowing how Tlr7 deletion benefits the learning process will help further dissect the mechanisms of cognitive impairment in AD brains and may prove useful for designing means of improving the learning defects of AD patients.
Among the various TLRs, TLR2 has been shown to recognize Aβ protein aggregates [16- 18]. Tlr2 deficiency increases Aβ aggregation and accelerates the cognitive decline in APP/PS1 transgenic mice [28]. Treatment of anti-TLR2 antibody for 7 months reduces microglial cell and astrocyte activation and improves spatial memory of APP/PS1 mice [36]. Thus, activation of the innate immune response has been suggested as a potential therapeutic approach for AD [28]. However, in our study, impairment of TLR7 activity by gene knockout has a beneficial effect on the spatial learning process of 5XFAD mice. Our data also indicates that Tlr7 deficiency does not influence Aβ deposits in 5XFAD brains (Figure 5 and 7a). Thus, although TLR7 and TLR2 are believed to use similar or at least overlapping signal pathways to initiate their downstream responses, the role of TLR7 in AD is different from that of TLR2. Thus, to develop potential therapeutic treatment of AD, the specificity of individual TLRs has to be taken into serious consideration to avoid negative side-effects.
Conclusions
We show here that although TLR7 may not regulate Aβ deposition and the activation of microglial cells and astrocytes in 5XFAD mice, removing TLR7 from 5XFAD mice specifically ameliorates the spatial learning process. Our results further reveal the diverse roles of various TLRs in AD.
Ethics approval and consent to participate
All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted. Human subjects were not involved in this work.
Consent to publication
Not applicable.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
H.-Y.L. and Y.-P.H. designed experiments; H.-Y.L., Y.-F.H., H.-R.L. and T.-L.Y. performed experiments; H.-Y.L. and Y.-F.H. analyzed data; H.-Y.L., Y.-F.H. and Y.-P.H. wrote the manuscript; Y.-P.H. secured financial support; all authors read the manuscript and approved the final version for submission.
Acknowledgements
We thank the Genomic Core Facility and Animal Facility of the Institute of Molecular Biology, Academia Sinica, for their assistance, members of Dr. Yi-Ping Hsueh’s laboratory for relabeling samples for blind experiments and technical assistance, and Dr. John O’Brien for English editing.
Funding
Y.-P. H. received grant supports from Academia Sinica (AS-103-TP-B05), the Ministry of Science and Technology (MOST 104-2321-B-001-050, 105-2321-B-001-031 and 105-2311-B-001- 061-MY3) and the Simons Foundation (SFARI# 388449). H.-Y. L. was supported by a fellowship from the Ministry of Science and Technology, Taiwan.
References
- Spangenberg EE, KN Green. Inflammation in Alzheimer's disease: Lessons learned from microglia-depletion models. Brain. Behav. Immun 61(1), 1-11 (2016).
- Hirano A, HM Zimmerman. Alzheimer's neurofibrillary changes. A topographic study. Arch. Neurol 7(1), 227-42 (1962).
- Kidd M. Paired helical filaments in electron microscopy of Alzheimer's disease. Nature 197(), 192-193 (1963).
- Miyakawa T, Uehara Y. Observations of amyloid angiopathy and senile plaques by the scanning electron microscope. Acta. Neuropathol 48(2), 153-156 (1979).
- Mandybur TI. The incidence of cerebral amyloid angiopathy in Alzheimer's disease. Neurology 25(2), 120-126 (1975).
- Cagnin A, DJ Brooks, AM Kennedy, et al. In-vivo measurement of activated microglia in dementia. Lancet 358(9280), 461-467 (2001).
- Fillit H, WH Ding, Buee L, et al. Elevated circulating tumor necrosis factor levels in Alzheimer's disease. Neurosci. Lett 129(2), 318-320 (1991).
- Dandrea MR, PA Reiser, NA Gumula, et al. Application of triple immunohistochemistry to characterize amyloid plaque-associated inflammation in brains with Alzheimer's disease. Biotech. Histochem 76(2), 97-106 (2001).
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol 11(5), 373-384 (2010).
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 140(6), 805-820 (2010).
- Zohaib A, Sarfraz A, Kaleem QM, et al. The Yin and Yang of Antiviral Innate Immunity in Central Nervous System. Curr. Pharm. Des 22(6), 648-655 (2016).
- Akira S. TLR signaling. Curr. Top. Microbiol. Immunol 311(1), 1-16 (2006).
- Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends. Immunol 33(9), 449-458 (2012)
- Dellacasagrande J. Ligands, cell-based models, and readouts required for Toll-like receptor action. Methods. Mol. Biol 517(1), 15-32 (2009).
- Park JS, Gamboni-Robertson F, He Q, et al. High mobility group box 1 protein interacts with multiple Toll-like receptors. Am. J. Physiol. Cell. Physiol 290(3), C917-24 (2006).
- Liu S, Liu Y, Hao W, et al. TLR2 is a primary receptor for Alzheimer's amyloid beta peptide to trigger neuroinflammatory activation. J. Immunol 188(3), 1098-1107 (2012).
- Chen K, Iribarren P, Hu J, et al. Activation of Toll-like receptor 2 on microglia promotes cell uptake of Alzheimer disease-associated amyloid beta peptide. J. Biol. Chem 281(6), 3651-3659 (2006).
- Jana M, Palencia CA, Pahan K. Fibrillar amyloid-beta peptides activate microglia via TLR2: implications for Alzheimer's disease. J. Immunol 181(10), 7254-7262 (2008).
- Tahara K, Kim HD, Jin JJ, et al. Role of toll-like receptor signalling in Abeta uptake and clearance. Brain 129(Pt 11), 3006-3019 (2006).
- Kariko K, Weissman D, Welsh FA. Inhibition of toll-like receptor and cytokine signaling-a unifying theme in ischemic tolerance. J. Cereb. Blood. Flow. Metab 24(11), 1288-1304 (2004).
- Green NM, Moody KS, Debatis M, et al. Activation of autoreactive B cells by endogenous TLR7 and TLR3 RNA ligands. J. Biol. Chem 287(47), 39789-39799 (2012).
- Cavassani KA, Ishii M, Wen H, et al. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J. Exp. Med 205(11), 2609-2621 (2008).
- Lehmann SM, Kruger C, Park B, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci 15(6), 827-835 (2012).
- Liu HY, Huang CM, Hung YF, et al. The microRNAs Let7c and miR21 are recognized by neuronal Toll-like receptor 7 to restrict dendritic growth of neurons. Exp. Neurol 269(1), 202-212 (2015).
- Oakley H, Cole SL, Logan S, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation. J. Neurosci 26(40), 10129-10140 (2006).
- Lund JM, Alexopoulou L, Sato A, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U S A 101(15), 5598-603 (2004).
- Lin CW, Hsueh YP. Sarm1, a neuronal inflammatory regulator, controls social interaction, associative memory and cognitive flexibility in mice. Brain. Behav. Immun 37(1), 142-151 (2014).
- Richard KL, Filali M, Prefontaine P, et al. Toll-like receptor 2 acts as a natural innate immune receptor to clear amyloid beta 1-42 and delay the cognitive decline in a mouse model of Alzheimer's disease. J. Neurosci 28(22), 5784-5793 (2008).
- Li Y, Du XF, Liu CS, et al. Reciprocal regulation between resting microglial dynamics and neuronal activity in vivo. Dev. Cell 23(6), 1189-1202 (2012).
- Signorino G, Mohammadi N, Patane F, et al. Role of Toll-like receptor 13 in innate immune recognition of group B streptococci. Infect. Immun 82(12), 5013-5022 (2014).
- Song W, Wang J, Han Z, et al. Structural basis for specific recognition of single-stranded RNA by Toll-like receptor 13. Nat. Struct. Mol. Biol 22(10), 782-787 (2015).
- Liu HY, Hong YF, Huang CM, et al. TLR7 negatively regulates dendrite outgrowth through the Myd88-c-Fos-IL-6 pathway. J. Neurosci 33(28), 11479-11473 (2013).
- Heneka MT, Kummer MP, Stutz A, et al. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493(7434), 674-678 (2013).
- Lehmann SM, Rosenberger K, Kruger C, et al. Extracellularly Delivered Single-Stranded Viral RNA Causes Neurodegeneration Dependent on TLR7. J. Immunol 189(3), 1448-1458 (2012).
- Liu HY, Chen CY, Hsueh YP. Innate immune responses regulate morphogenesis and degeneration: roles of Toll-like receptors and Sarm1 in neurons. Neurosci. Bull 30(4), 645-54 (2014).
- McDonald CL, Hennessy E, Rubio-Araiz A, et al. Inhibiting TLR2 activation attenuates amyloid accumulation and glial activation in a mouse model of Alzheimer's disease. Brain. Behav. Immun 58(1), 191-200 (2016).